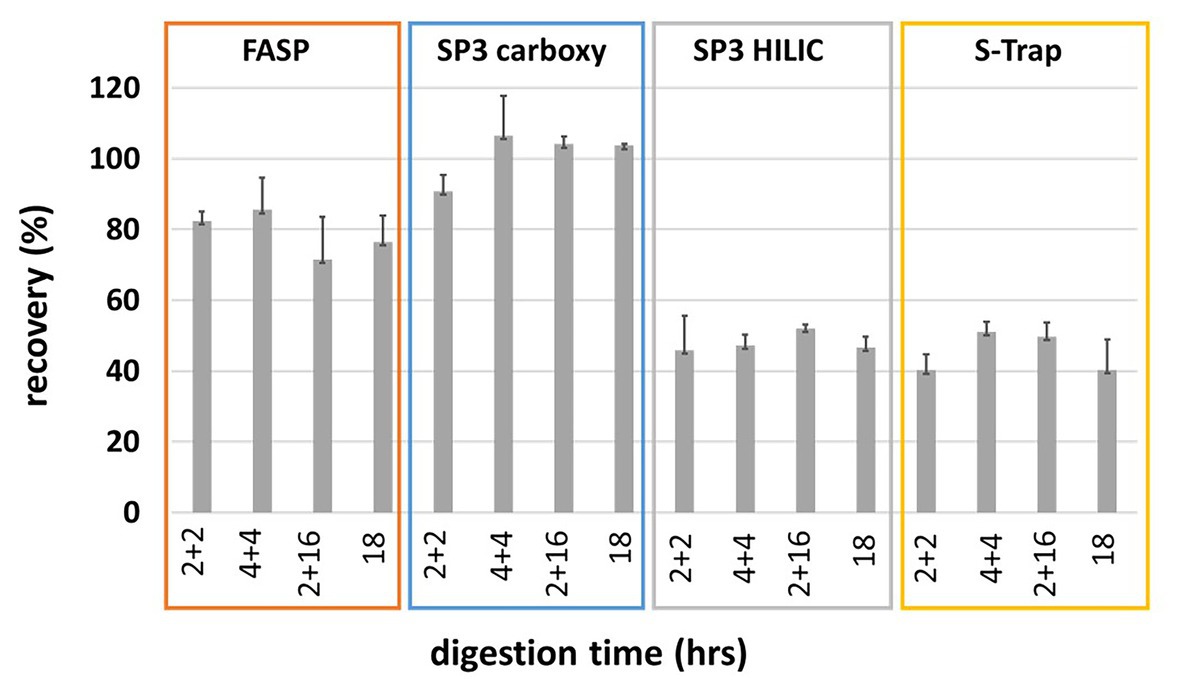
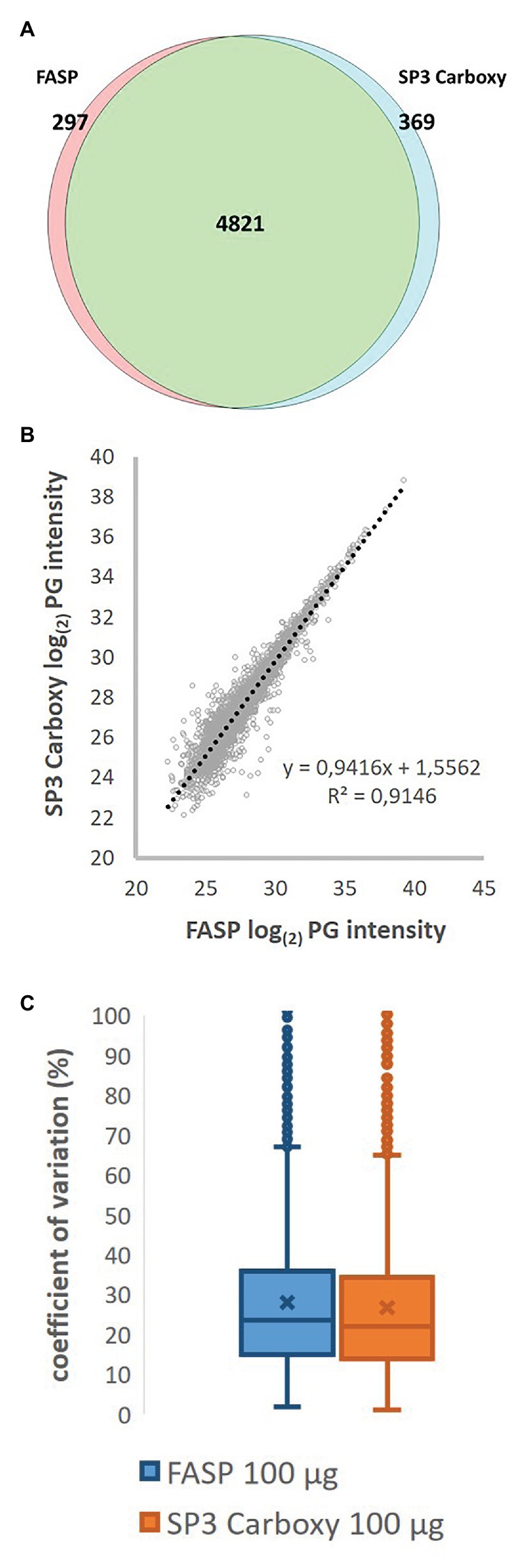
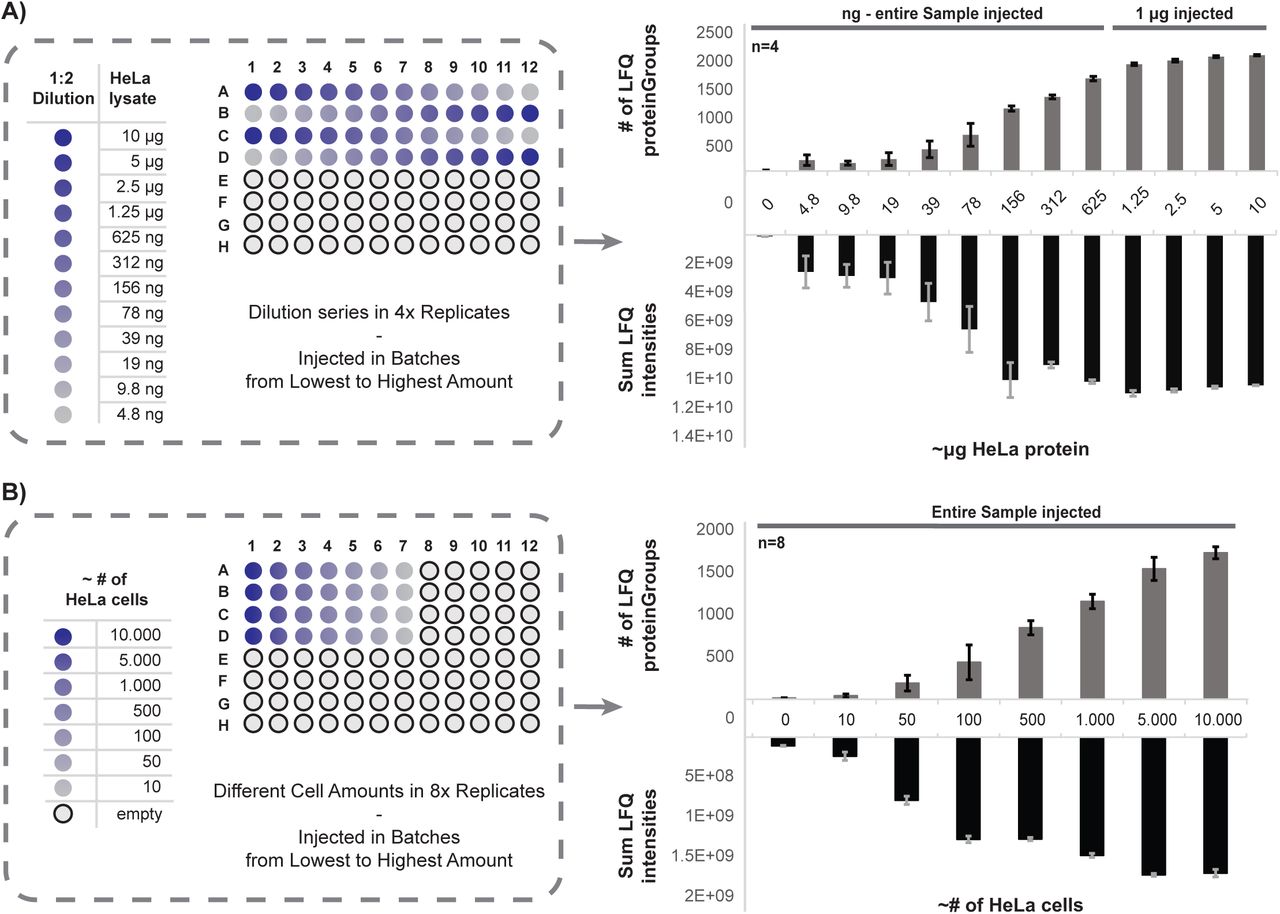
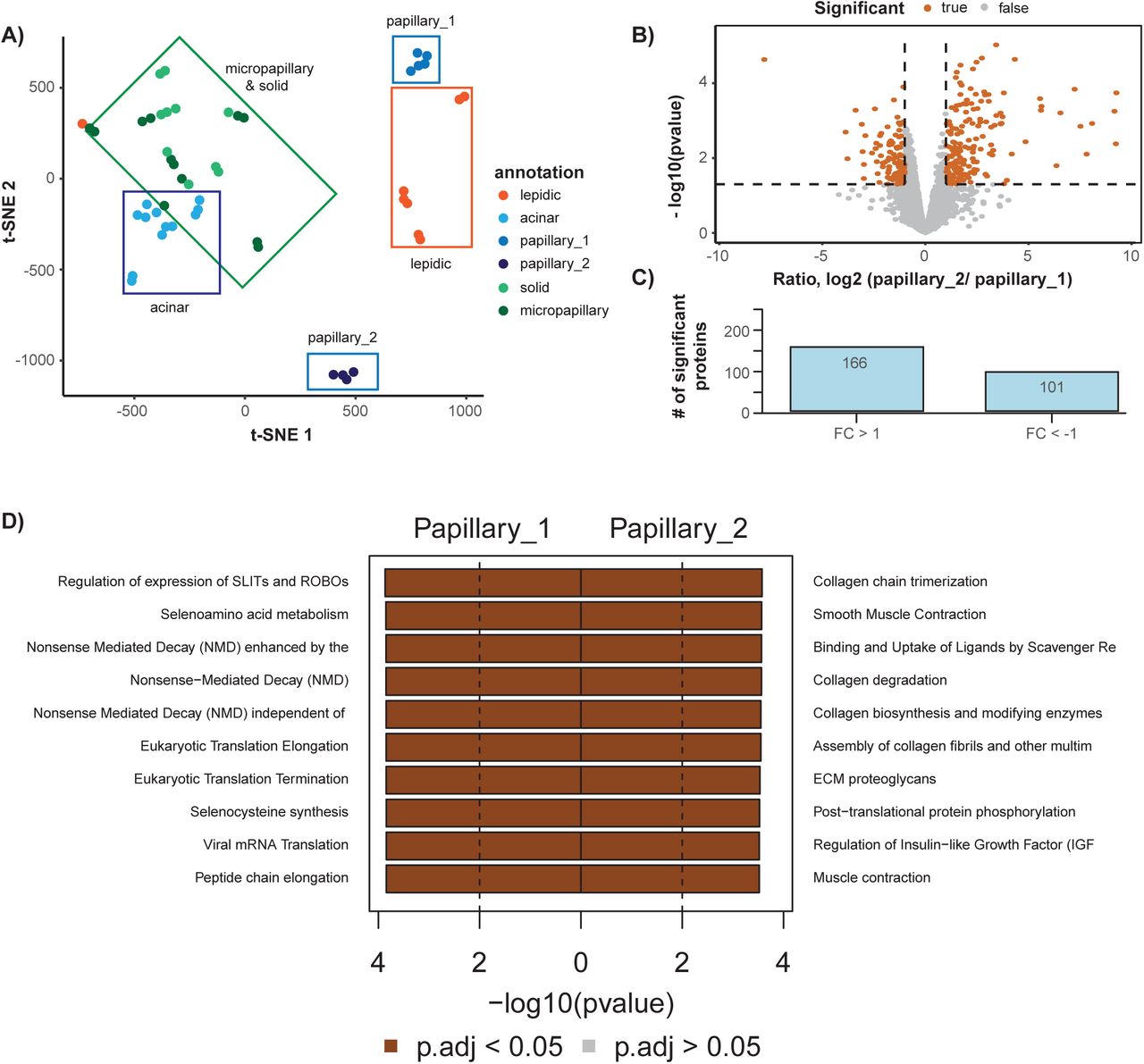
Modular automated bottom-up proteomic sample preparation for high-throughput applications | PLOS ONE
Modular automated bottom-up proteomic sample preparation for high-throughput applications | PLOS ONE

SCASP: A Simple and Robust SDS-Aided Sample Preparation Method for Proteomic Research - Molecular & Cellular Proteomics
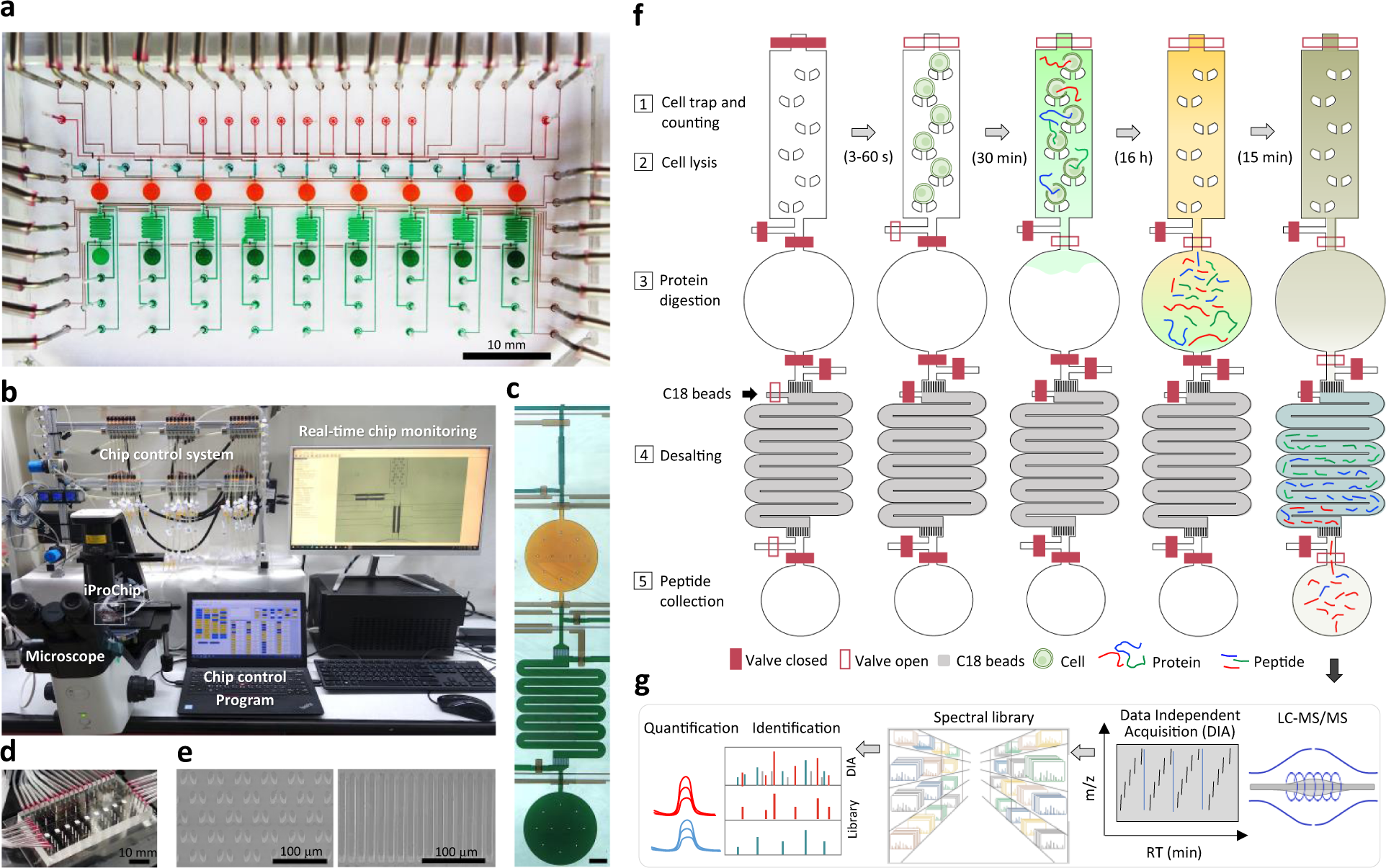
Streamlined single-cell proteomics by an integrated microfluidic chip and data-independent acquisition mass spectrometry | Nature Communications

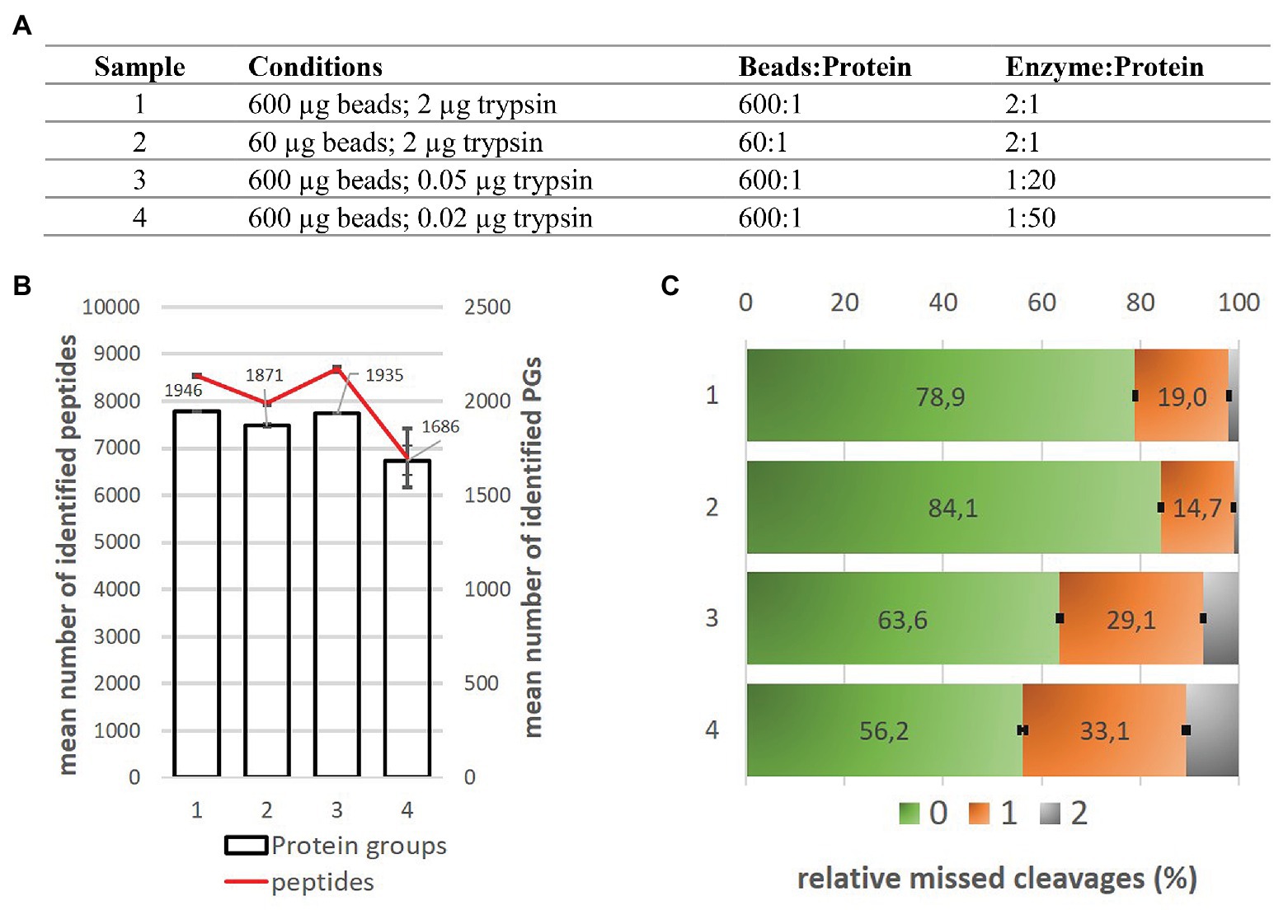
Automated proteomic sample preparation: The key component for high throughput and quantitative mass spectrometry analysis - Fu - - Mass Spectrometry Reviews - Wiley Online Library

SP3-Enabled Rapid and High Coverage Chemoproteomic Identification of Cell-State–Dependent Redox-Sensitive Cysteines - Molecular & Cellular Proteomics

Extending the Compatibility of the SP3 Paramagnetic Bead Processing Approach for Proteomics | Journal of Proteome Research
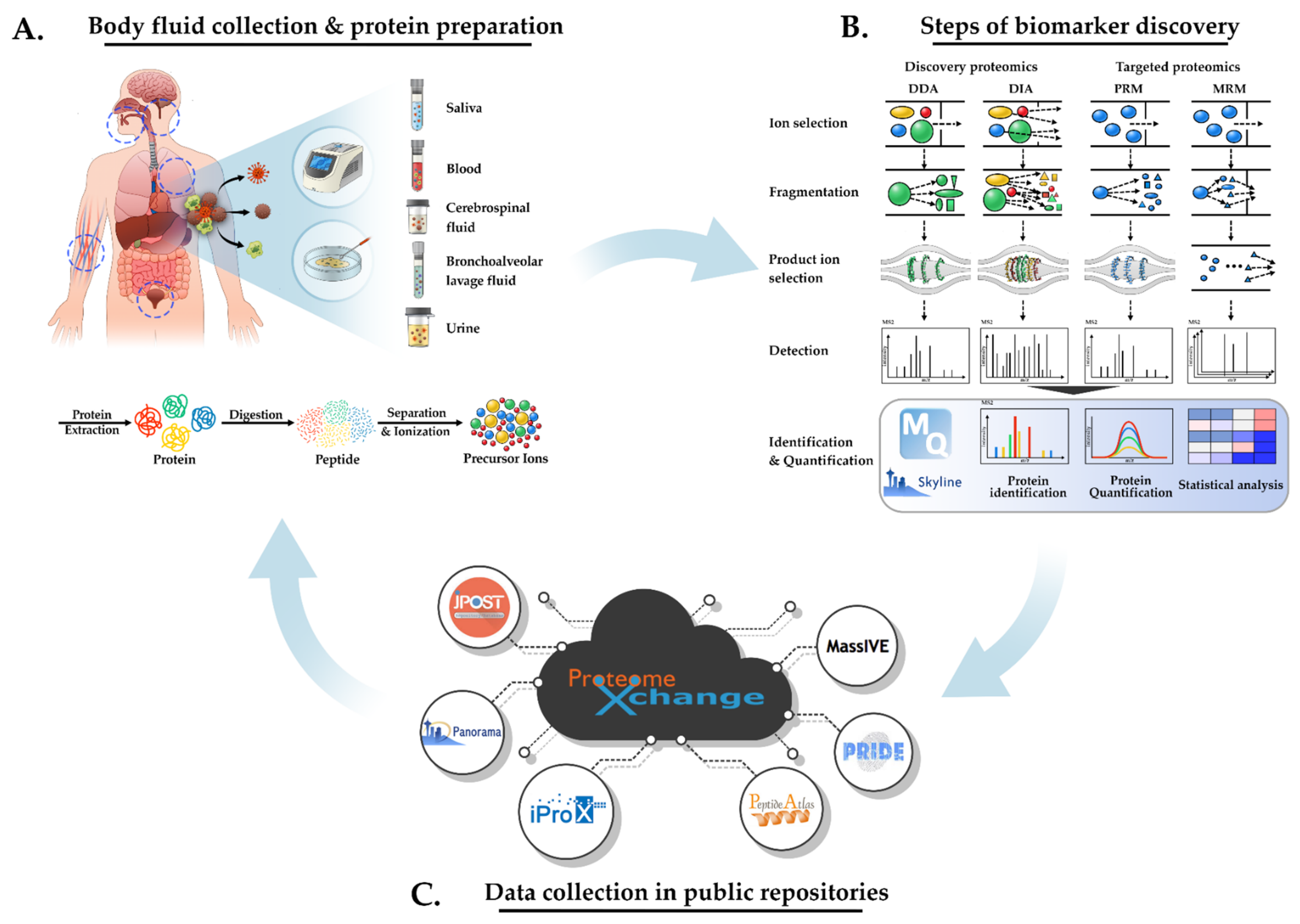
IJMS | Free Full-Text | Review of Liquid Chromatography-Mass Spectrometry-Based Proteomic Analyses of Body Fluids to Diagnose Infectious Diseases | HTML
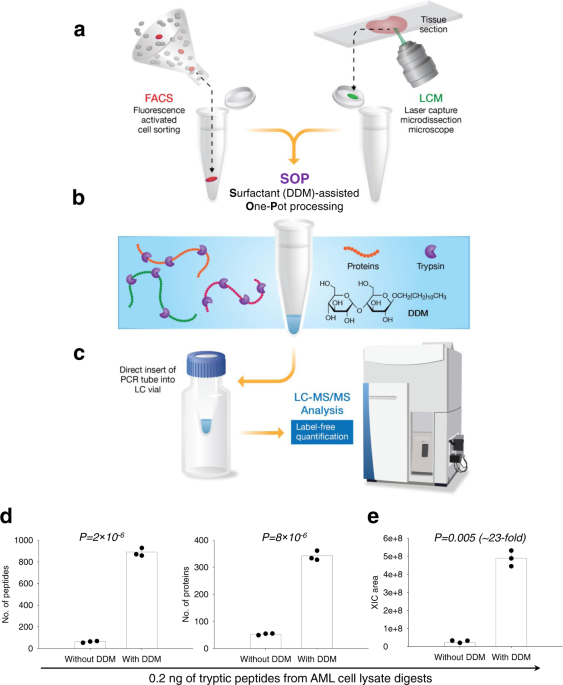
Surfactant-assisted one-pot sample preparation for label-free single-cell proteomics | Communications Biology
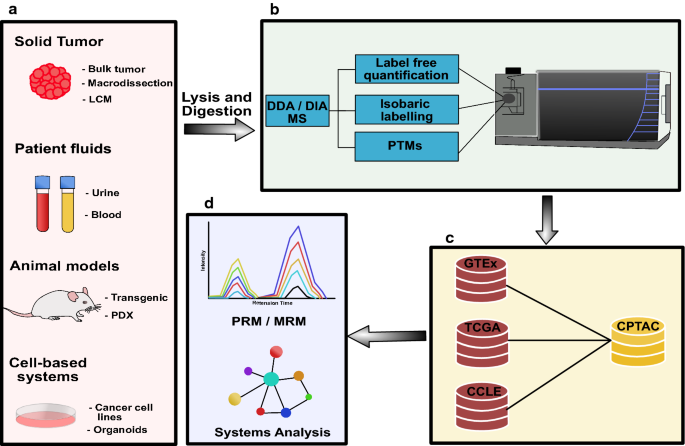
Recent advances in mass spectrometry based clinical proteomics: applications to cancer research | Clinical Proteomics | Full Text